Constrained bonds¶
A typical molecular system often contains different modes of motion at very different timescales. Of all these motions, bond vibrations are some of the fastest, compared with, say, the rotational motion of the molecules.
Molecular dynamics calculations must therefore use a timestep that is small enough in order to adequately track the fastest mode of motion in the system. Otherwise, too large a timestep can lead to instability and inaccuracy of motion.
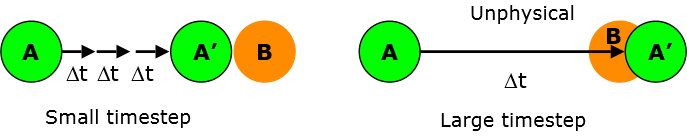
Bonds that are connected to lighter atoms tend to vibrate faster, especially those that contained the hydrogen atoms. For instance, consider a typical C-H vibrational bond, which has a typical frequency of 4000 cm-1 . This corresponds to a period of ~8 fs (femtoseconds). It means the timestep used to update the atomic positions must be much less than the period of the bond vibration and this is usually in the range of 0.5-1.0 fs.
Note
You can see why it might not be feasible to use MD simulations to look at long timescale phenomena. For example, to model atomic motions spanning across 1 ns by using a 1 fs timestep would require 1,000,000 MD iterations.
However, the timestep can be increased provided the MD simulation does not need to track bond vibration. If you do not need very accurate thermodynamic values, removing bond vibrations does not contribute significantly to the overall results. This can be achieved by constraining bonds to a fixed equilibrium length by using a constraint algorithm, e.g. SHAKE.
The SHAKE algorithm basically involves two stages: (1) determining the positions of atoms in the absence of bond constraints, and (2) determining the deviation in the length of a given rigid bond, to calculate retrospectively the constraint force (directed along the bond) needed to conserve the bond length. This process will be repeated if the largest deviation is found to exceed the desire tolerance with respect to the fixed bond length.
Note
There is a trade off in terms of computational time, depending on how precisely you want to fix the bond lengths. The higher precision you require, the more SHAKE iteration steps would be needed, which will take up more computational resources.
In DL_POLY, the tolerence limit is set to 10-6 and the maximum SHAKE iteration step is set to 250 by default. These values can be changed in the DL_POLY CONTROL file. For example, to reduce the tolerance to 10-7 and the maximum number of iterations per timestep to 100, older versions of DL_POLY can use the following directives:
mxshak 100
tolerance 1.0e-7
while DL_POLY_5 uses the following:
shake_max_iter 100
shake_tolerance 1.0e-7
If bonds that contain the hydrogen atoms are constrained, then the timestep can be increased to 2 fs. For soft matters and biological molecules such as proteins, the tolerance can be reduced to around 10-4 .