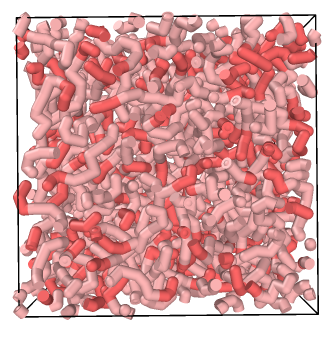
Visualising Simulation¶
The results from a DL_MESO_DPD simulation can include particle trajectories in the HISTORY file. These can be visualised by loading the data into a particle visualisation software package, e.g.
The trajectory data in the HISTORY file needs to be converted to file formats that either package can read. Utilities included with DL_MESO can be used, which can be compiled according to the instructions in the Ready, get set, go guide. Alternatively, Python scripts are available to convert the HISTORY file into a variety of formats, all of which require a module to read in HISTORY files (dlmhistoryread.py). These scripts also require the Python modules docopt and tqdm to be installed using a command such as:
pip install docopt tqdm
| TIP: | Some Python 3 installations may use python3 in place of python and pip3 instead of pip. |
|---|
VMD:
To convert the HISTORY file for VMD, either use the traject_vtf.exe utility in the same directory, e.g.
./traject_vtf.exe
or use the history_dlm_to_vtf.py Python script along with the dlmhistoryread.py module
python history_dlm_to_vtf.py
Both the utility and the script will generate a traject.vtf file in VMD’s VTF trajectory format, which contains the particle coordinates for all available trajectory frames and (if included) a list of particle pairs connected together by bonds. If any molecules are included in the original HISTORY file, these are identified as residues in the VTF file.
This file can now be opened in VMD, which should automatically detect it as the correct file type. Options are available in VMD to change the representations of particles and/or bonds, including selecting which particle species or molecules/residues are visible. A playback bar is available to ‘playback’ the simulation as an animation, which can be exported to a video file.
| TIP: | If no bond list is included in a VTF file (e.g. for a system without molecules), VMD will try to add bonds between particles based on how apart they are from each other. Since DPD particles are likely to be quite close together, VMD will report errors in its terminal window stating several particles have too many bonds, but this will not stop VMD from loading and visualising the file and those messages can therefore safely be ignored. |
|---|
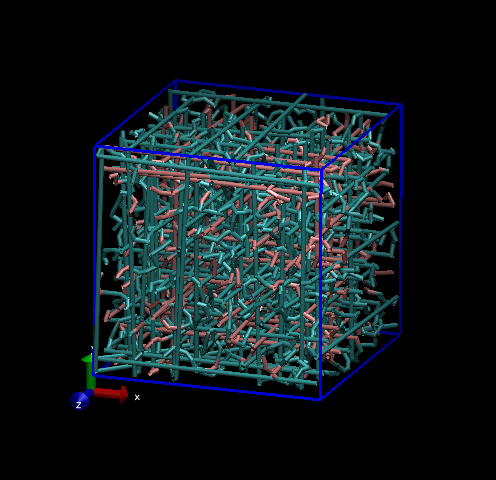
If your simulation includes bonds that stretch across the box due to periodic boundary conditions, it is possible to make VMD move segments of molecules beyond the bounds of the box to give minimum bond lengths. Find the terminal window opened by VMD and type:
pbc join res -first first -last last
to join up the molecules (residues) in all available frames. An example of this for the polymer melt can be seen below.

VMD can also read the following other file formats, most of which do not include bond connectivity data but can be generated from DL_MESO_DPD HISTORY files using Python scripts:
- CHARMM DCD files (
history_dlm_to_dcd.py) - DL_MESO/DL_POLY CONFIG files (
history_dlm_to_config.py) - each consisting of a single frame/configuration - DL_POLY HISTORY files (
history_dlm_to_history_dlp.py)
DCD files also require a PSF (protein structure file) to identify particle species/molecule types and provide bond connectivity information, which can be generated using the history_dlm_to_psf.py script and loaded into VMD with the DCD file.
OVITO:
To convert the HISTORY file for OVITO, either use the traject_xml.exe utility in the same directory, e.g.
./traject_xml.exe
or use the history_dlm_to_galamost_xml.py Python script along with the dlmhistoryread.py module
python3 history_dlm_to_galamost_xml.py
Both the utility and the script will generate a series of files in an XML-based format for GALAMOST (a GPU-based molecular simulation code) numbered according to the trajectory frame number. Each file contains the particle coordinates, velocities for all particles (if provided in the HISTORY file) and a list of particle pairs connected together by bonds for each frame. (Note that the bond connectivity data does not change during a DL_MESO_DPD simulation.)
These files can be opened in OVITO by selecting either a range of them or just the first one. Options are available to change how the particles, bonds and simulation cell are displayed. A playback bar exists to ‘playback’ the simulation as an animation, which can be exported to a video file.
Unlike VMD, you do not need to make adjustments for bonds that stretch across the periodic box in OVITO: these are automatically cut for individual trajectory frames, as shown below for the polymer melt example.