Force field schemes¶
Due to the classical nature of force fields, there are a variety of ways to derive potential parameters: these also use different functions to model the systems. Each research group tends to stick to certain fitting procedures and functions to establish their FF models, hence the birth of different FF schemes.
The diagram below shows a section of a FF family tree indicating how different schemes were born and evolved.
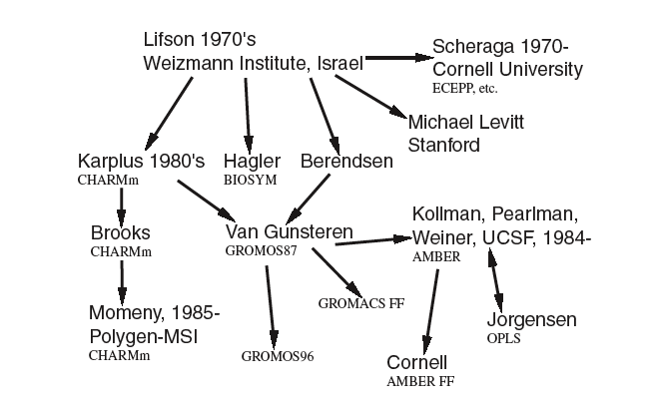
A new scheme was usually formed by borrowing some of the characteristics from previous schemes; this was sometimes done to model new or different classes of materials. This is the reason why two different FF schemes can sometimes share similar sets of potentials and, more often, the same functional forms.
However, the tree has been growing steadily since the 1970s and has never stopped. New branches are constantly added and some have even started with a completely new origin and different versions to map new classes of materials or to improve upon the qualities of a previous FF when modelling the same class of materials.

FF types
An FF scheme is usually constructed with the aim to use it in a specific manner, which means it can perform better for certain materials than others.
The qualities of a FF can generally be divided into two types:
Specific FF schemes. These are better suited to model certain specific types of materials. For instance, proteins, DNA, carbohydrates, molecular cages, ionic liquids, etc. These are usually fitted with many parameters and the accuracy is generally high for the right type of materials.
However, transferability to model other molecules may still be acceptable as these molecules or their derivatives were often formed as part of the fitting criteria during the construction of the scheme.
Generic FF schemes. These schemes can have fewer parameters but are applicable to a wide range of molecule types and can be used to model general classes of molecules. For this reason, they have wide applicability, although the accuracy may be poorer.
Note
Some modern FF schemes can be pretty sophisticated compared with their predecessors and the applicability of these FF schemes can straddle across both of the above types. Some of these schemes can be used to model general small drug molecules quite accurately with the parameters fitted to tens of thousands of test molecules.

Example FF schemes
The list below shows some popular FF schemes for biomolecules and organic molecules. Note that the list and their applicabilities are by no means exhaustive. It should be used as a rough guide only.
- CHARMM (Chemistry at HARvard using Molecular Mechanics) - biomolecules
- AMBER (Assisted Model Building with Energy Refinement) - biomolecules
- OPLS (Optimized Parameters for Large-scale Simulations) - general organic molecules in condensed phases
- MMFF (Merck Molecular Force Field) - small drug molecules
- MM2, MM3, MM4 - Allinger et al. - organic molecules
- UFF (Universal Force Field) - general molecules, metallic molecules, most elements across the Periodic Table
There are also other variations of the above mentioned schemes. For example:
- AMBER Glycam - for carbohydrates
- CHARMM CGenFF, AMBER GAFF - for general molecules such as small drug molecules
- OPLS for ionic liquids
- etc.