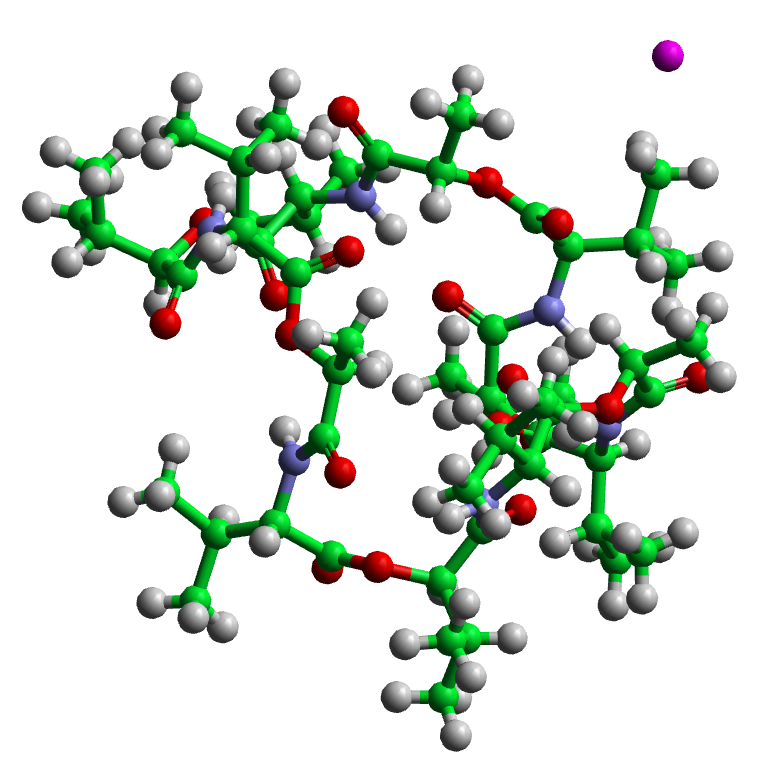
Potassium Capture by Valinomycin¶
Summary¶
This exercise explores the ability of a naturally occurring molecule to chelate the potassium ion and is an example of the application of molecular simulation to the study of biological activity. The study demonstrates the use of simulation to reveal the relationship between the structural properties of a molecule and its biological function.
Background¶
Valinomycin is a relatively small, naturally occurring, cyclic molecule comprised of 168 atoms. It strongly resembles a peptide with 12 α −amino acids, except that half of the amide bonds, NH − CO, are replaced by ester linkages, O − CO, and there are changes in optical stereochemistry at some of the α -carbons (technically it is a cyclic dodecapsipeptide), see the above figure. The molecule demonstrates a remarkable flexibility, but its most significant property is its ability to wrap around a potassium ion (shown in purple), thus giving the ion a hydrophobic coat which permits its transfer through the hydrophobic membranes of living cells. Valinomycin is thus a molecule essential to life.
This study cannot hope to address all the properties of this remarkable molecule, and the student is warned against reading too much into the results of the experiments, which neglect many of the contributing factors to the full biological processes, but it will hopefully give some clues as to how the molecule functions in the way it does. The simulations will attempt the capture of a potassium ion by a valinomycin molecule in vacuo, ref. Capture of potassium ions by valinomycin: a molecular dynamics simulation study
If successful, the simulations should reveal some of the molecular features that facilitate the capture of potassium in real life.
Task¶
- Download the files:
FIELD,CONTROL,CONFIGand display the system using CONFIG. Note the structural features: the CO groups of the ester and amide links, which have a role in the chelation of the potassium; the methyl and isopropyl groups, which form the hydrophobic coat; and features that may give rise to hydrogen bonding within the molecule itself and help stabilise its various conformational forms. - Take a look at the FIELD file and see how the intramolecular terms of the Valinomycin are specified: bonds, valence angle and dihedral angle potentials are described. These are based on the AMBER forcefield. It should give you some idea about how to specify more complex molecules.
- At the bottom of the CONFIG file the position of the potassium ion is defined. It is some distance away from the valinomycin, the centre of mass of which is close to the origin. Run a simulation starting with this CONFIG file. What you are looking for is an indication that the valinomycin recognises the presence of the potassium ion and draws it closer.
- Consider why a neutral molecule like valinomycin should have an affinity for a charged species like potassium. What structural features are responsible? Is the molecule responding in the way you expect (for example conformationally)?
- If things are happening too slowly, you may consider changing the formal charge on the potassium, just to see how the system responds.
- Repeat the simulation with the potassium starting in different positions. Is there a directional preference?
- Run a simulation and create a HISTORY file. Write a simple routine to calculate the dipole moment of the valinomycin at intervals and see if this correlates with the potassium position.
A sample video of the simulation can be seen on youtube at: Potassium Capture by Valinomycin